|
I am a Ph.D. student in the Department of Computer Science at Virginia Tech. My advisor is Dr. Debswapna Bhattacharya. I am interested in computational biology, applied machine learning, data science and bioinformatics, optimization, and human-computer interaction. Currently, my research is primarily focused on macro-molecular predictive modeling, with a specific emphasis on intra- and inter-molecular interaction prediction using cutting-edge artificial intelligence techniques. I am a recipient of the Pratt Fellowship Award, 2023 at Virginia Tech. Prior to starting my Ph.D., I completed my Master of Science in Computer Science and Software Engineering at Auburn Univerisity (2021), and my Bachelor of Science in Computer Science and Engineering at Bangladesh University of Engineering and Technology (BUET) (2016). Google Scholar / LinkedIN / Email / CV / twitter |

|
|
|
|
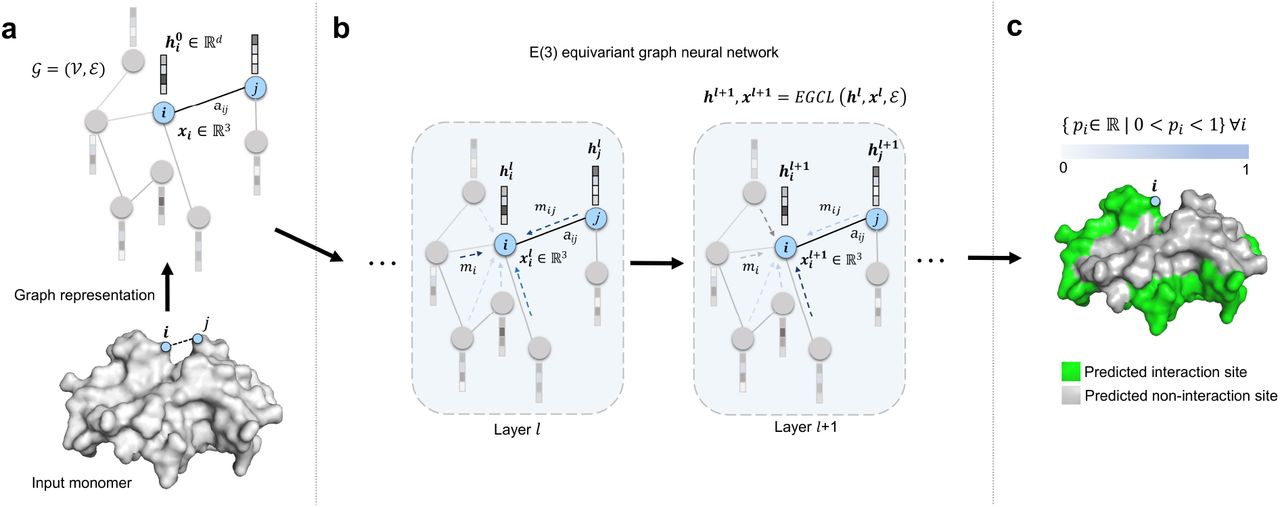
Rahmatullah Roche, Bernard Moussad, Md Hossain Shuvo and Debswapna Bhattacharya PLOS Computational Biology, 2023 GitHub A robust and accurate protein-protein interaction site prediction method leveraging symmetry-aware graph convolutions with equivariant transformation of translation, rotation, and reflection in 3D space. |
|
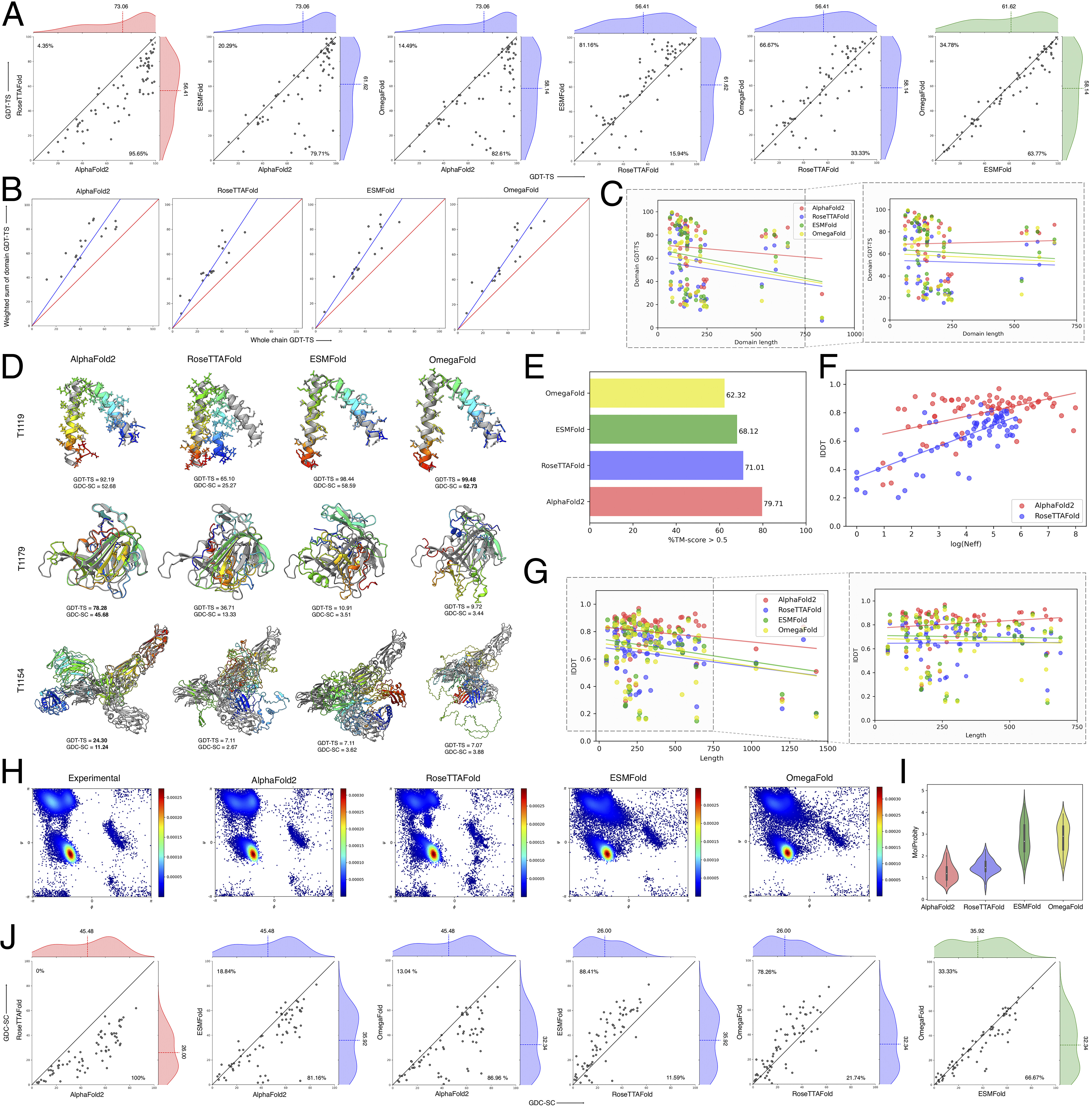
Bernard Moussad, Rahmatullah Roche and Debswapna Bhattacharya Proceedings of the National Academy of Sciences, , 2023 GitHub This benchmarking showcases the power of transformers revolutionizing protein structure prediction, and points towards future improvements. |
|
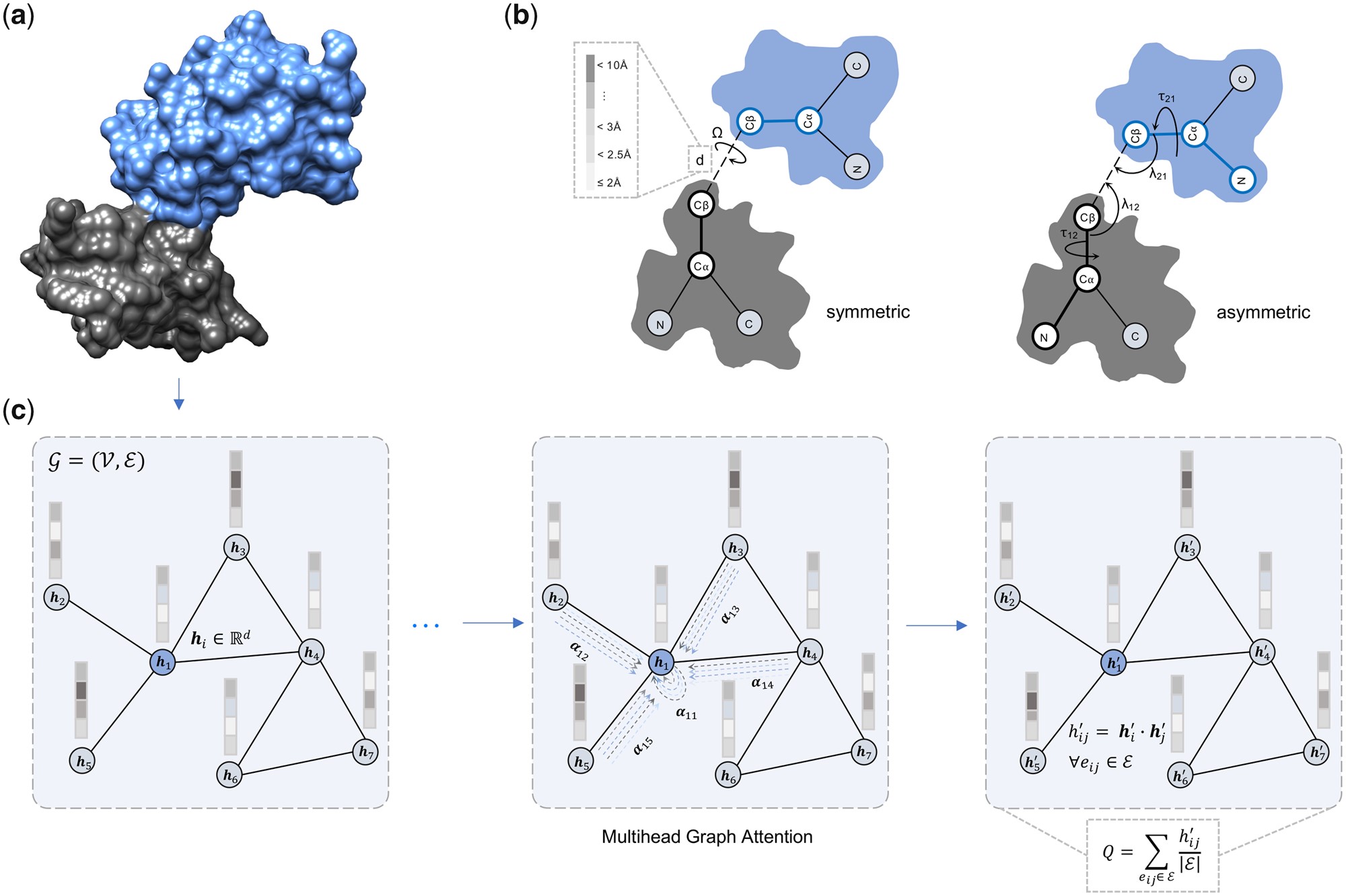
Md Hossain Shuvo, Mohimenul Karim, Rahmatullah Roche and Debswapna Bhattacharya Bioinformatics Advances, , 2023 GitHub A deep graph learning-based method utilizing a multi-head graph attention network for accurate protein-protein interface quality estimation. |
|
Sutanu Bhattacharya, Rahmatullah Roche, Md Hossain Shuvo, Bernard Moussad and Debswapna Bhattacharya Homology Modeling: Methods and Protocols, Methods in Molecular Biology, 2023 This book chapter presents an overview of contact-assisted threading methods, discussing recent advances, current limitations, and future prospects for their application in enhancing the accuracy of low-homology protein modeling. |
|
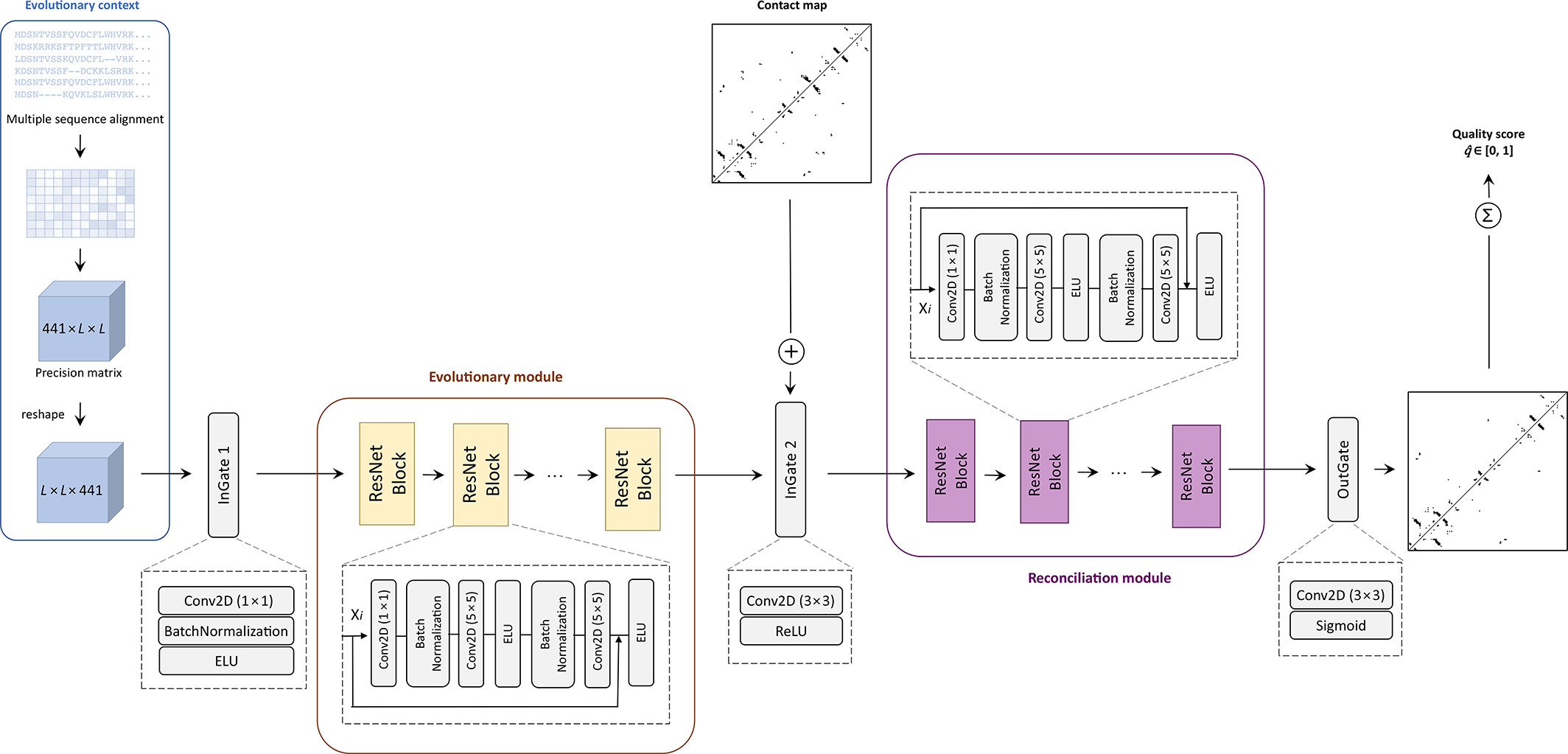
Rahmatullah Roche, Sutanu Bhattacharya, Md Hossain Shuvo and Debswapna Bhattacharya PROTEINS: Structure, Function, and Bioinformatics, 2022 GitHub A deep learning method that assesses the quality of protein contact maps with high accuracy, leveraging evolutionary context and residue-pair consistency. |
|
|
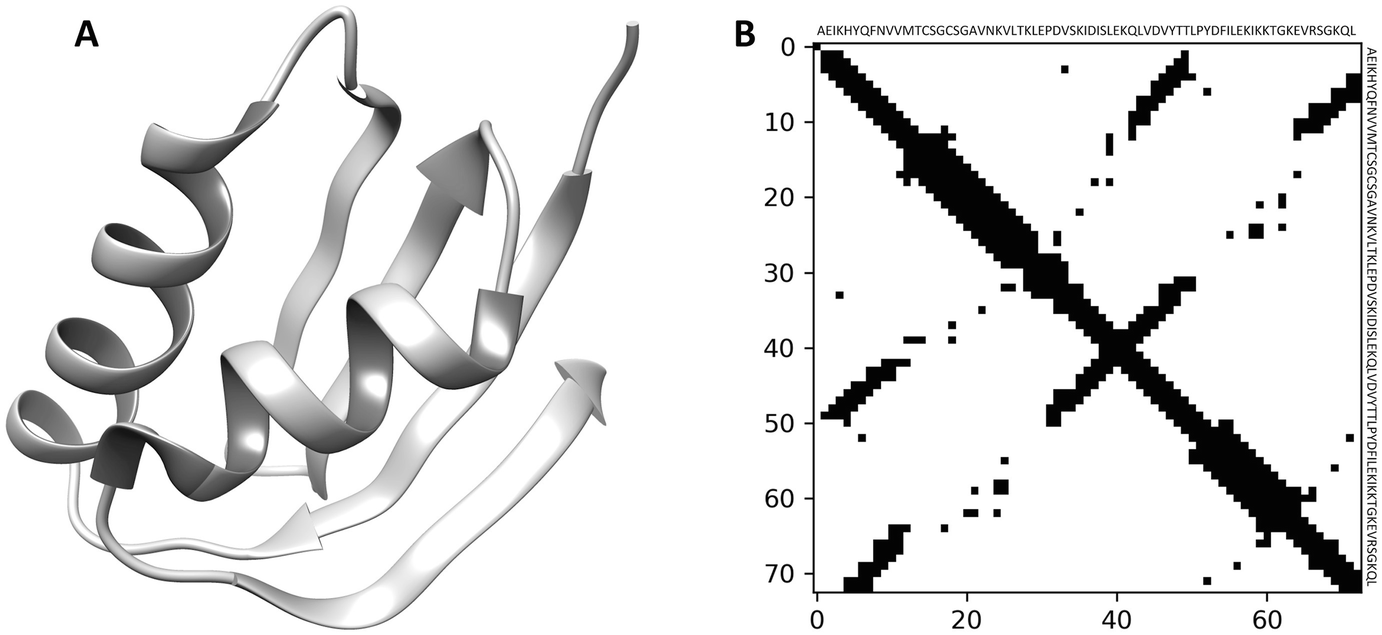
Rahmatullah Roche, Sutanu Bhattacharya and Debswapna Bhattacharya PLOS Computational Biology, 2021 GitHub DConStruct is a hierarchical structure modeling method that uses inter-residue interaction maps of varying resolutions to enhance ab initio protein folding. Our method demonstrats improved folding accuracy for soluble and membrane proteins compared to existing approaches, overcoming the limitations of experimental structure determination protocols. |
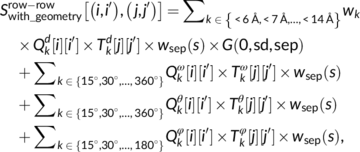
|
Sutanu Bhattacharya, Rahmatullah Roche, Bernard Moussad and Debswapna Bhattacharya PROTEINS: Structure, Function, and Bioinformatics, 2021 GitHub A deep learning method that assesses the quality of protein contact maps with high accuracy, leveraging evolutionary context and residue-pair consistency. |
|
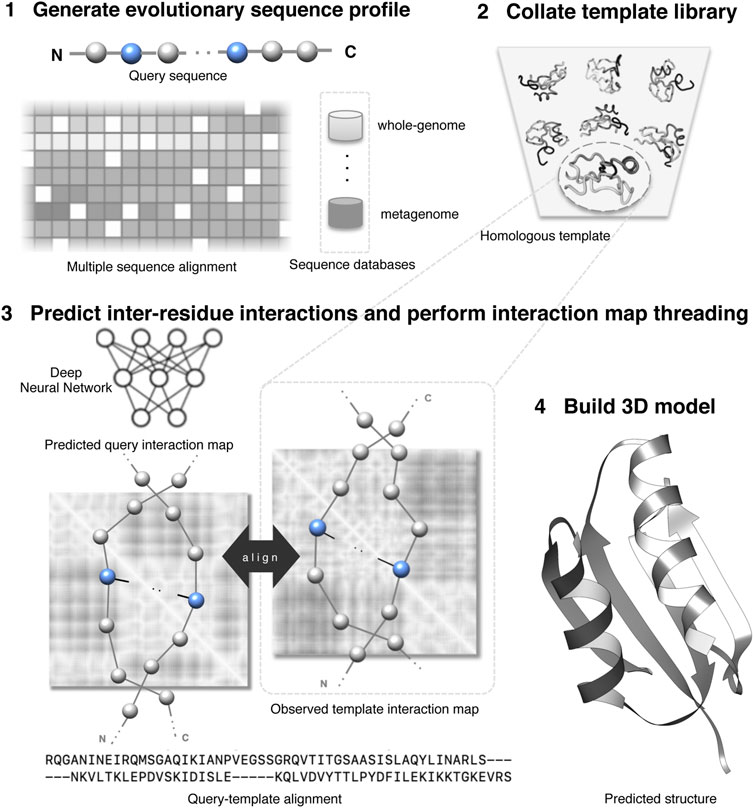
Sutanu Bhattacharya, Rahmatullah Roche, Md Hossain Shuvo and Debswapna Bhattacharya Frontiers in Molecular Biosciences, 2021 This review paper highlights the emerging trends in distant-homology protein threading using predicted interaction maps and discusses avenues for enhancing sensitivity in homology detection. |
|
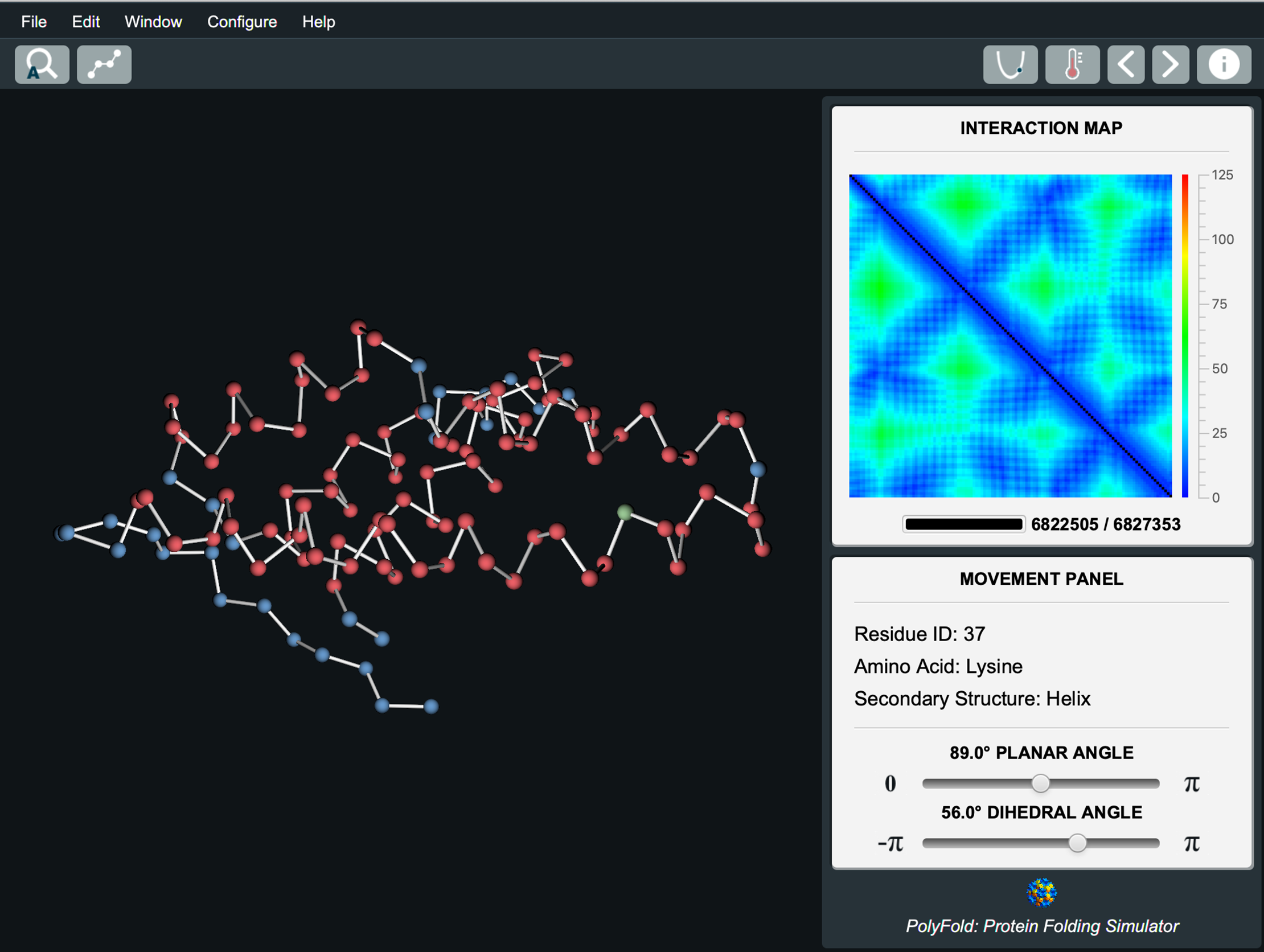
Andrew J. McGehee, Sutanu Bhattacharya, Rahmatullah Roche, Bernard Moussad and Debswapna Bhattacharya Plos One, 2020 GitHub PolyFold, an open-source interactive simulation tool, offers intuitive protein folding visualization for citizen science and scientific exploration. |
|
R. Roche, S. Bhattacharya, D. Bhattacharya, “Hybridized distance- and contact-based hierarchical structure modeling for folding soluble and membrane proteins,”
in
BCB '21: Proceedings of the 12th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics , USA, August 2021
|
|
|
|
|
|
|