
Stochastic Modeling of Budding Yeast Cell Cycle
This work is a collaboration with Dr. Tyson's Lab .
We combine the model builder JigCell with the
stochastic simulation toolkit StochKit . This translation enables us to do stochastic simulation for the budding yeast cell cycle model .
Here is a visualization tool we built to demonstrate the dynamics of our model: Budding Yeast Cell Cycle Visualization

Motivation
During the past decade, many successful deterministic models of macromolecular regulatory networks
have been built. Deterministic simulations of these models can show only average dynamics of the systems.
However, stochastic simulations of macromolecular regulatory models can account for behaviors that are
introduced by the noisy nature of the systems but not revealed by deterministic simulations.
In this research project, we apply state-of-the-art stochastic simulation techniques to significant macromolecular regulatory models.
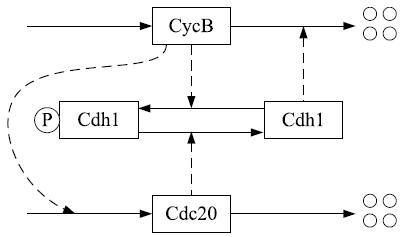
Network diagram of a simple regulatory network
Methods
Since there currently exist models of value, it is natural to wish to run the existing models using stochastic simulation
algorithms, even if those models were created with continuous, deterministic simulators in mind. Our vision
is that in practice modelers will develop initial models using deterministic ODEs for their relative speed and
simplicity, before progressing to a full stochastic simulation.
Unfortunately, ODE-based models are not cast in the right form for direct simulation using stochastic
methods. The values of species in ODE models are usually written in concentration form.
However, stochastic simulations require the model to
be in terms of population because they account for individual molecules. Thus, a conversion process must
take place. Moreover, the values in deterministic models are often normalized to avoid the trouble of
defining real concentrations. When converting the values to number of molecules, however,
efforts must be taken to scale normalized values back to real values.
For large and complex models that could have over one hundred parameters, this process would
be tedious and error prone to perform by hand.
We have built an automatic conversion tool in our JigCell model builder to ease this process.
Our emphasis on units of values in the model allows converting large and complex models manageable.
After directly converting the model from the deterministic form to the stochastic form, we added
various features that are unique to stochastic simulations, such as growing volume and random cell division.
Our finally derived model of budding yeast cell cycle is the most complex and reallistic stochastic model
on cell cycle to date. An advanced stochastic simulation package StochKit, which implements Gillespie's Stochastic Simulation Algorithm
and its variations, are used to perform stochastic simulations on the model.
By doing simulations on this model, we obtained some biologically significant and interesting results.
For example, we found some mutants are on the edge of being alive or dead, i.e., they have certain probability to cease division.
Deterministic simulations are not able to accurately capture this feature because they always give binary answers. Using stochastic simulations,
however, we obtained results that are more conforming to the real world data.
The three figures below showcase how stochastic simulations capture the behavior of a half-dead mutant,
which is not able to be captured by deterministic simulations.
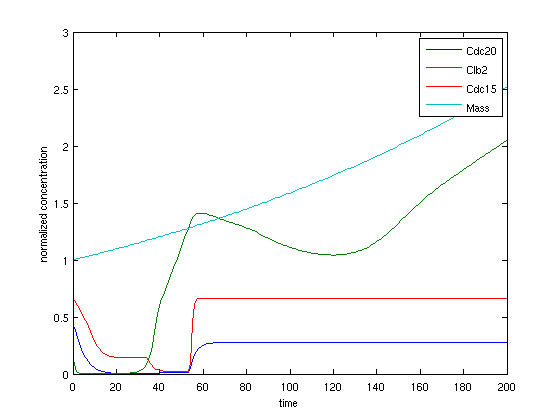
Deterministic simulation result. The cell always died without any division.
.png)
One instance of stochastic simulation. The cell underwent one division and continued to grow.
.png)
Another instance of stochastic simulation. The cell died without any division.
Stochastic simulations are known for being computationally intensive, and thus require high performance
computing facilities to be practical. We used parallel computing on Virginia Tech's supercomputer
System X to obtain statistical data of our realistic cell cycle model.


.png)
.png)