Multiscale Modeling of Bacterial Chemotaxis with Interactive User Interface
Introduction
Chemotaxis has been a well studied chemical phenomenon for bacteria. To illustrate this kind of interesting behaviors, many models have been built. However, most of them did not have visualizations. In the year of 2005, using StochSim, a more complete simulator AgentCell was introduced, with the inclusion of the alternate moving patterns of the bacterium, and graphic display of the bacteria moving in an environment with a simple distribution of attractant chemicals.
Recently, Bray's another new model, using their existing BCT program, made a success in providing matching behaviors of the bacteria with the experimental data observed in lab, by introducing infectivity to the model for achieving the high sensitvity of the bacterium cell for the chemical, and by increasing the adaptation rate. However, this model so far still deals with simple environments.
As we realized, it would be very interesting and useful to see the bacteria reacting to more complicated and realistic maps of attrant or repellent chemicals, visualized by a more user-friendly application interface. This project focuses on developing a graphic interface, which enables the diffusion of the chemicals, allows users to configure the whole environment map, and represents the visualization of the moving bacteria. Furthermore, it should provide at least three different walking mechanisms for the purpose of comparison. In the future, this interface is also expected to model chemotaxis phenomena with population of bacteria. We believe in the value of our interface both in the field of research and education.
Chemotaxis
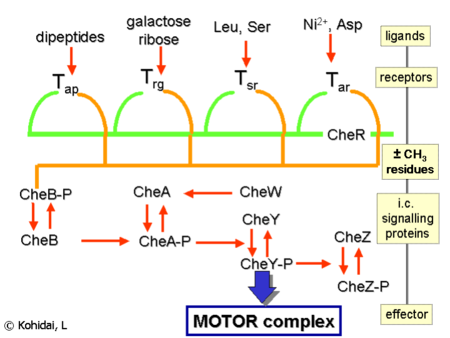
It has been discovered that bacteria swim towards the more highly concentrated area of attractant chemicals and avoid environments with higher repellent concentrations. This kind of behavior is called chemotaxis. Briefly speaking, becteria achieve this by sensing chemical concentrations outside the cell with their receptors, then inducing a series of chemical reactions in the cell, and finally deciding where to go, by changing their swimming patterns.
A complicated chemical network describes how signals such as attractants or repellents outside the cell are transducted through the pathway and affect the moving patterns of the cell.

According to observations in experiments, bacteria such as E-coli can respond to very tiny change of concentration of attractant. This kind of sensitivity is another interesting phenomenon and it has far exceeded the sensitivity of existing computer models. In Bray's recent model, they assume that the binding of one receptor by attractant chemical can induce its neighbor receptors to act as if they were bound by the same chemical as well, thus amplifying the sensitivity of the model. They call this mechanism the infectivity and it provided matching data with experiments.
Overview of the Interface
Currently, our interface can be mainly divided into two parts, the simulation environment map and the operation panel. The map is two-dimensional and has a 100x100 resolution thus making 10,000 grids. On the operation panel the user can select different kinds of chemicals and their concentration, and paint them onto the map.
We have noticed that, previous models can only work on a quite limited number of scenarios of environments. In most cases, they simply deal with environments with a constant chemical concentration over the whole map, or a environment map divided into two parts, one of which has no chemicals and the other half having a constant concentration. However these are far from a realistic environment which should be much more complicated but meaningful. For this consideration, our interface allows users to configure the map. The user can set areas with different concentrations of chemicals and see the response of the bacteria.
Also our interface provides three kinds of walking strategies for the bacteria. The purpose of this is for comparison. The three strategies include random walk mode, run/tumble mode and the real chemical model mode. When simulation starts, bacteria under different mechanisms will move on the map specified by the user from the initial position until the user pause the simulation.
Right now, we are focusing on improving the model to best match the experimental data, especially in how to integrate the real network into the model. Then, more complicated phenonmena may be considered for simulation.