 N.Jiang et al.: A System Model of the Insulin Secretion Network
N.Jiang et al.: A System Model of the Insulin Secretion NetworkMultiscale Modeling, Simulation, and Sensitivity Analysis of Biochemical Systems Motivated by Pulsatile Insulin Secretion
The Goal
The overall goal of the project is to develop innovative techniques for a modeling, simulation and analysis framework for biochemical systems and this framework should be able to support dynamic, automatic switching between different models and algorithms with convenient deterministic and stochastic sensivity analysis tools to analyze the robustness of systems of interest.
Introduction
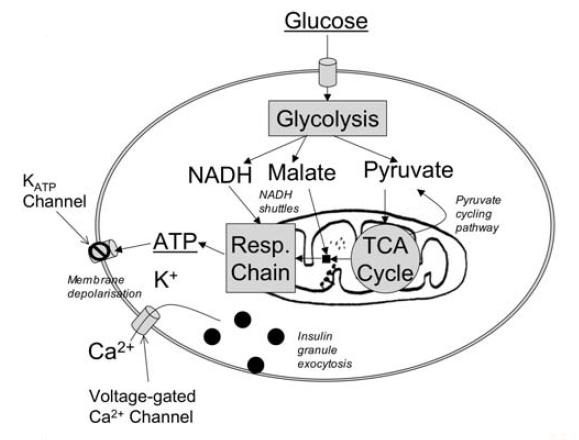
To test this modeling, simulation and analysis framework, it will be applied to study the mechanism of coupled oscillations in calcium ion (Ca2+) and cellular ATP/ADP driven by mitochondria, which is directly related to the defect in pulsatile insulin secretion in diabetes. As we know, Diabetes Mellitus is a leading cause of death in the developed world. Currently 25-30 percent of adults have a high at risk of becoming diabetic. In healthy individuals insulin is secreted into the bloodstream after a meal as a signal for muscles and fat to use the glucose produced from the digested food. This signaling process maintains low blood glucose levels. In diabetes this signaling process is disrupted. The oscillations of insulin concentration become abnormal. The main pathways involved in glucose-stimulated insulin secretion is shown in the following figure. Insulin is contained in vesicles just below the plasma membrane of pancreatic beta cells. Three signals are required for the fusion of these vesicles with the plasma membrane to release insulin into the blood. These signals are ATP, calcium (Ca2+), and amino acids made from mitochondrial TCA cycle intermediates. All three of these signals are controlled primarily by mitochondria. Therefore, to model and simulate mechanism of coupled oscillations in calcium ion and cellular ATP/ADP driven by mitochondria can better help biologists understand the pulsatile insulin secretion in diabetes at different levels, aiding the development of future clinical treatments.
 N.Jiang et al.: A System Model of the Insulin Secretion Network
N.Jiang et al.: A System Model of the Insulin Secretion Network
Methods
We propose to develop a multiscale computational model of coupled oscillations in calcium ion and cellular ATP/ADP driven by mitochondria. The oscillation mechanism is studied at different scales. First, we will build a model of the oscillation in a single mitochnodrian, Once the initial model of oscillation in an individual mitochondrion is validated, the spatial and coupling effects will be introduced to model and simulate the coupled oscillations among 100 to 200 mitochondria in a single cell. Next, massively parallel computation will be applied to simulate the coupled oscillations among 1,000 to 10,000 beta cells that make up a single Islet of Langerhans in the pancreas. The robustness of the oscillation mechanism to parameter variation and spatial heterogeneity will be studied at all scales, with tools developed for hybrid (deterministic and stochasitc) sensitivity analysis.
Significance
This project rigorously tests the hypothesis that synchronization of the oscillations of the multiple mitochondria in each beta cell and between beta cells is necessary for the correct secretion of insulin. It will also help us gain further insight into the mechanisms involved in the coupled oscillation of multiple cells and the robustness of insulin release mechanism, and provide a modeling, simulation and analysis framework for similar biological systems.