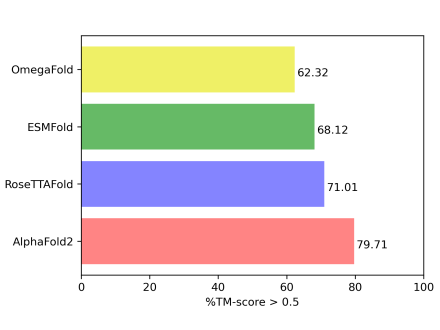
I am an Associate Professor in the Department of Computer Science at Virginia Tech, based in the Blacksburg campus. My primary research interests are computational biology & bioinformatics (CBB) and applied machine learning (ML) with a particular focus on artificial intelligence (AI) for structural biology. I am also interested in AI-driven biomedical data science for the extraction of knowledge from massive biological datasets.
The long-term goal of my research is to deepen our understanding of the fundamental biomolecular sequence-structure-function relationship through the lens of computational modeling. My current research takes a critical step toward this goal by developing, evaluating, and disseminating a new generation of knowledge-guided and integrative machine learning methods and resources for modeling and characterization of biomolecules at scale.
Prospective lab members: If you are interested in joining my group at Virginia Tech, please read this.
BIO
I obtained my PhD in Computer Science from the University of Missouri - Columbia in 2016 under the supervision of Prof. Jianlin Cheng. Before joining Virginia Tech in 2022, I was an Assistant Professor at Auburn University from 2017 to 2021. Selected honors and awards that I am a recipient of include:
- National Science Foundation National AI Research Resource Award, 2024
- National Distinction and Outstanding Contributor, Virginia Tech, 2023
- National Science Foundation CAREER Award, 2020
- Ginn Faculty Fellowship, Auburn University, 2020
- National Institutes of Health Maximizing Investigators' Research Award (MIRA) for ESI, 2020
- Outstanding Engineering Faculty Award, Auburn University, 2019
- Outstanding CS PhD Student Award, University of Missouri-Columbia, 2014
RESEARCH
Active Projects:
Current Group Members:
- Xingyue Feng: PhD Student in CS at VT (2025 - Present)
- Sriniketh Vangaru: Undergraduate Student in CS at VT (2024 - Present)
- Xinyu Wang: PhD Student in CS at VT (2024 - Present)
- Sumit Tarafder: PhD Student in CS at VT (2022 - Present)
Former Group Members:
- Trevor Norton: Postdoc at VT (2023 - 2025). Now Senior Data Scientist at RTX Corporation.
- Rahmatullah Roche: PhD in CS at VT (2018 - 2024). Now Assistant Professor at Columbus State University.
- Md Hossain Shuvo: PhD in CS at VT (2018 - 2023). Now Assistant Professor at Prairie View A&M University.
- Sutanu Bhattacharya: PhD in CSSE at AU (2017 - 2021). Now Assistant Professor at Auburn University Montgomery.
- Rahul Alapati: MS Thesis in CSSE at AU (2017 - 2019). Now Software Engineer at Microsoft.
- Andrew J. McGehee: BS in CSSE at AU (2018 - 2020). Then Software Engineer at Google.
- Carmen Stowe: BS in CSSE at AU (2017 - 2018). Then MIT Lincoln Laboratory and MS Student at Georgia Tech.
Visitors and Interns:
- Maryam Haghani: Graduate Student in CS at VT co-advised with Prof. T. M. Murali (2023 - 2025)
- Erin Wienke: Graduate Student in CS at VT (2024)
- Nabayan Chaudhury: Graduate Student in CS at VT (2023 - 2024)
- Bernard Moussad: Graduate Student in CS at VT (2021 - 2024)
- Sajib Acharjee Dip: Graduate Student in CS at VT (2023)
- Mohimenul Karim: Graduate Student in CS at VT (2021 - 2023)
- Fatemeh Sarshartehrani: Graduate Student in CS at VT (2022)
- Muhammad Gulfam: Graduate Student in CSSE at AU (2020 - 2021)
- Hudson Chromy: Undergraduate Student in CSSE at AU (2019)
- Behnam Rasoolian: Graduate Student in CSSE at AU (2017 - 2018)
- Tian Xia: Graduate Student in CSSE at AU (2017)
- Donald Holyfield: Undergraduate Student in CSSE at AU (2018)
- Swati Baskiyar: Undergraduate Student in CSSE at AU (2018)
Note to Prospective Students and Postdocs:
I am always looking for self-motivated students and postdocs to join my group. Unfortunately, I am not able to respond to all emails. So, depending on your situation, please follow one of the follwing routes:- You are a prospective PhD student: please apply directly to CS at VT PhD program and list me as a faculty of interest. Afterwards, send me an email with the subject line "[Prospect] Interested in joining your group (PhD)" with a CV and a brief description of your interests. Admissions are centralized and decisions are made by committee.
- You are a student already at VT: send me an email with the subject line "[Prospect] Interested in joining your group (VT Student)" with a CV and a brief description of your interests and background.
- You are a prospective Postdoc: send me an email with the subject line "[Prospect] Interested in joining your group (Postdoc)" with a CV, link to your publications, and a brief description of your interests and background.
TEACHING
Past, Current, and Upcoming Courses:
- Fall 2025: CS 5805. Advanced Machine Learning (VT)
- Fall 2024: CS 4824. Machine Learning (VT)
- Fall 2024: CS 6824. AI-powered Molecular Modeling (VT)
- Spring 2024: CS 4824. Machine Learning (VT)
- Spring 2022: CS 6824. AI-powered Molecular Modeling (VT)
- Spring 2022: CS 4824. Machine Learning (VT)
- Fall 2021: COMP 3270. Introduction to Algorithms (AU)
- Spring 2020: COMP 5960. Computational Biology (AU)
- Fall 2020: COMP 3270. Introduction to Algorithms (AU)
- Spring 2019: COMP 5960. Computational Biology (AU)
- Fall 2018: COMP 3270. Introduction to Algorithms (AU)
- Spring 2018: COMP 5960. Computational Biology (AU)
- Fall 2017: COMP 3270. Introduction to Algorithms (AU)
- Spring 2017: CS 898AR. Machine Learning in Biology (WSU)
- Fall 2016: CS 797I. Introduction to Bioinformatics (WSU)
RESOURCES
Extramural Funding:
Active
Active
Completed